A common task in data analysis is the need to find specific patterns within text data. Pattern searching and matching play a crucial role in filtering, transforming, and extracting vital information.
In R, this is carried out with the help of the grep function.
The grep function in R helps users identify character patterns within a string vector and returns matching indices.
Another related function, grepl, provides a logical vector to indicate pattern matches with a vector.
Together, these functions enable users to efficiently filter, subset, and transform their data based on specific criteria, providing greater control and flexibility in handling complex datasets.

In this article, we will cover the grep function in detail. We’ll explore its purpose, functionality, and syntax, and talk about some related functions.
We will also look at several examples, work with regular expressions, handle UTF-8 strings, and much more.
By the end, you should have a solid understanding of how the grep function works and how you can use it for data analysis.
So, get ready to upgrade your pattern-matching skills in R.
Understanding Grep Function in R
The main function of grep() is to search for matches of a given pattern within each element of a character vector.
This is particularly useful for filtering and processing data, especially when it comes to text analysis or data-cleaning tasks.

Here are some key aspects of the grep function:
- It searches for matches to an argument pattern within the given character vector.
- It returns a vector of the indices of the input vector elements where matches are found.
- The grepl() function, a related function, returns a logical vector indicating whether a match was found TRUE or not FALSE.
In the latter part of this section we will go over the following:
- Syntax of Grep
- Function Parameters of Grep
- A Simple Example of Gerp
1) What is The Syntax of Grep
Here’s the basic syntax for using the grep() function in R:
grep(pattern, x, ignore.case = FALSE, fixed = FALSE, value = FALSE)2) What are The Function Parameters of Gerp
To modify the behavior and output of the grep() function, you can adjust the following arguments:
- pattern: Character string to match against the specified elements of the string.
- x: The specified string vector to search for the pattern within.
- ignore.case: A boolean setting, either TRUE or FALSE, to indicate whether the search should be case-sensitive or not.
- By default, this is set to FALSE.
- fixed: A boolean setting, either TRUE or FALSE, that defines if the pattern should be treated as a fixed string or a regular expression.
- When fixed = TRUE, the pattern is treated as a literal string without any special characters. By default, this is set to FALSE.
- value: A boolean setting, either TRUE or FALSE, determining whether to return the matched elements themselves (TRUE) or their indices (FALSE). By default, this is set to FALSE.
3. A Simple Example of Gerp
Consider the following scenario as an example.
Suppose you have a list of passenger names stored in a vector passengers and you wish to find the index of any passenger with the first name ‘David’.
By using the grep function, you can easily do this.
Here’s the code:
grep("David", passengers)The output will look something like this:

How to Use Grep With Character Strings and Regular Expressions
The grep function in R offers a sophisticated and flexible way to perform pattern-matching tasks, whether it’s matching simple character strings or dealing with more complex regular expressions.

Let’s explore some common scenarios and how grep can be effectively utilized. We will go over the following:
- Matching a Character String
- Working With Regular Expressions
- Matching With Multiple Patterns
1) Matching a Character String
When working with character strings, grep() can be used to identify any substrings that match a specific pattern within a vector or a vector of character strings.
Here’s an example R script to show the basic use of grep:
# Create a character vector
words <- c("apple", "banana", "cherry", "date", "fig", "grape")
# Use grep() to find words that contain "a"
grep("a", words)The code is looking for the indices of the elements in the words vector that contain the letter “a”.
This is referred to as partial matching.
Here is the output:

Here’s a breakdown of how grep() arrived at the result:
- “apple” contains the letter “a”, so index 1 is included.
- “banana” contains the letter “a”, so index 2 is included.
- “cherry” does not contain the letter “a”, so index 3 is not included.
- “date” contains the letter “a”, so index 4 is included.
- “fig” does not contain the letter “a”, so index 5 is not included.
- “grape” contains the letter “a”, so index 6 is included.
2) Working With Regular Expressions
Regular expressions are a powerful tool for pattern matching and data extraction in R.
They can be used with the grep() function to search for more complex patterns within character strings.
To use regular expressions in grep(), make sure to set the parameter fixed = FALSE (the default value) or perl = TRUE.
Here’s an example:
# Create a vector 'words' with different fruit names in it
words <- c("apple", "banana", "cherry", "date", "fig", "grape")
# Use grep() with a regular expression to find words that start with a vowel and end with "e"
grep("^[aeiou].*e$", words)Here’s a breakdown of the regular expression used:
- ^: This asserts the position at the start of a line.
- [aeiou]: This matches any one of the vowels “a”, “e”, “i”, “o”, or “u”.
- .*: This matches any character (except a newline) 0 or more times.
- e$: This matches the character “e” at the end of a line.
This should return only the index 1, indicating that “apple” matches the given regular expression.
Here’s the output in RStudio:

3) Matching With Multiple Patterns
Matching with multiple patterns can be achieved in R by using the grep and grepl functions with various pattern options:
- Fixed Patterns
- Multiple Patterns
- Position-based matching
1) Fixed Patterns
By setting the fixed = TRUE argument in grep or grepl, you can search for an exact matching substring within each element of a character vector.
This can be useful when working with a large dataset and specific text patterns need to be found.
2) Multiple Patterns
Using the pipe symbol | in your perl style regular expressions allows you to match multiple patterns.
For example, grep(“pattern1|pattern2”, character_vector) matches elements containing either “pattern1” or “pattern2”.
3) Position-based Matching
Functions like startsWith() can be used to only match patterns at the beginning of a character string.
This can help reduce false matches and improve processing efficiency.
What is The Alternative to the Grep Function?
The grepl function in R is similar to the grep function but serves a slightly different purpose.
Specifically, grepl searches for matches of a particular pattern within a vector and returns a logical vector indicating whether each element of the vector contains the given pattern.

The basic syntax for using grepl is as follows:
grepl("pattern", x)Here is a simple example demonstrating the use of grepl:
# Sample character vector
text <- c("apple", "banana", "grape", "orange", "pineapple")
# Search for the pattern "ap"
pattern_match <- grepl("ap", as.character(text))
# Output: (TRUE, FALSE, FALSE, FALSE, TRUE)
print(pattern_match)The above code gives the following output:

What is The Difference Between Grep and Grepl
While both grep and grepl are used to search for patterns within a character vector, they differ in their output and functionality.
grep:
- Returns a vector of indices of the character strings where matches were found.
- This function is useful when you need the position of the matches within the vector.
grepl:
- Returns a logical vector (consisting of TRUE and FALSE)
- Indicates whether each element of the character vector contains the given pattern.
- Useful when you want a quick binary (yes or no) answer for each vector element.
Here is a side-by-side comparison using the same example as before:
# Sample character vector
text <- c("apple", "banana", "grape", "orange", "pineapple")
# Grep
grep("ap", as.character(text))
# Output: (1, 5)
# Grepl
grepl("ap", as.character(text))
# Output: (TRUE, FALSE, FALSE, FALSE, TRUE)Here’s the output:

Grep With Dataframes
The grep() function is particularly useful when there’s a need to select rows or columns in an R dataframe based on specific text patterns.
Here are the steps to filter rows based on patterns in the stored data:
- Define the Pattern to Match: Choose the specific text pattern you’re looking for.
- Use grep() to Find Matching Rows: Apply the grep() function to the desired column, specifying the pattern. The value argument controls whether the matching elements or their indices are returned; in this context, set it to FALSE to get the indices.
- Subset the Data Frame: Use the indices obtained from grep() to subset the original data frame and create a new one containing only the matching rows.
The following example shows how to apply these steps:
# Sample data frame
data_frame <- data.frame(
column_1 = c("keyword here", "other text", "another keyword"),
other_column = c(10, 20, 30)
)
# Pattern specification
pattern <- "keyword"
# Use grep() to find indices of rows with the pattern in the "column_1" column
filtered_rows <- grep(pattern, data_frame$column_1, value = FALSE)
# Subset the data frame using the filtered row indices
new_data_frame <- data_frame[filtered_rows, ]
new_data_frameHere’s the output:

The grep() function isn’t limited to filtering rows, either. It can also be applied to search for and select columns in a data frame based on a specific text pattern.
Here’s a step-by-step example to showcase this functionality:
- Define the Pattern to Match: Identify the specific text pattern you want to find in the column names.
- Use grep() to Find Matching Columns: Apply the grep() function to the column names of the data frame, specifying the pattern. Setting the value argument to TRUE will return the matching column names.
- Subset the Data Frame: Use the column names having the matched pattern to subset the original data frame and create a new one containing only those columns.
Here’s the code that puts these steps into action:
# Sample data frame
data_frame <- data.frame(
variable_A = c(10, 20, 30),
variable_B = c(40, 50, 60),
other_column = c(70, 80, 90)
)
# Pattern specification
pattern <- "variable"
# Use grep() to find column names that match the pattern
matching_columns <- grep(pattern, colnames(data_frame), value = TRUE)
# Subset the data frame using the matching column names
selected_columns_data_frame <- data_frame[, matching_columns]
selected_columns_data_frameAnd, here’s the output of the above code:

Additional Pattern-Matching Functions in R
R offers several other pattern-matching functions that allow for more complex and detailed matching:
- sub: Used to replace the first occurrence of a pattern in a given character vector with a specified replacement value.
- gsub: Similar to sub, but gsub replaces all the matching occurrences of a pattern in a given character vector with a specified replacement value.
- regexpr: Used for regular expression matching and returns the position of only the first occurrence of a pattern in each element of a character vector.
- gregexpr: Similar to regexpr, but gregexpr returns the positions and lengths of all occurrences of a pattern in each element of a character vector.
- grepRaw: Similar to grep, but used for matching raw vectors.
- regexec: Returns the positions and lengths of capturing groups within the pattern matches for each element of a character vector.
- regmatches: Used for extracting matched substrings based on the results obtained from the functions regexpr, gregexpr, and regexec.
- agrep: Similar to grep, but allows for approximate matching, meaning that it can find matches that are close to the given pattern.
Working with Encodings and Locales
Pattern matching within text data often involves dealing with various encodings and locales.
The grep function in R offers functionality that can handle both single-byte and multibyte character sets, catering to different languages and scripts.

1) Handling Single-Byte and Multibyte Locales
In R, the grep function allows for pattern matching with character vectors and works with different encodings and locales.
The default regular expression engine in R uses the current single-byte locale suitable for ASCII-only matching, but it can also handle multibyte locales when necessary.
Using Multibyte Locales
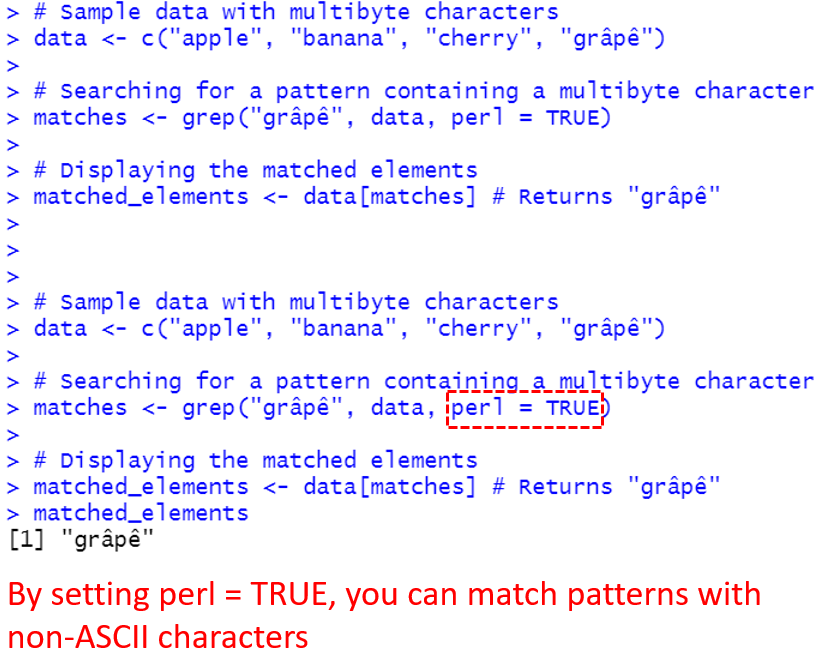
If you need to work with multibyte characters (such as those found in some non-English alphabets), you can enable Perl-compatible regular expressions by setting perl = TRUE.
# Sample data with multibyte characters
data <- c("apple", "banana", "cherry", "grâpê")
# Searching for a pattern containing a multibyte character
matches <- grep("grâpê", data, perl = TRUE)
# Displaying the matched elements
matched_elements <- data[matches] # Returns "grâpê"
matched_elements The above code gives the following output:
Optimizing Performance With useBytes:
To enhance performance and avoid potential issues with resource limits, you can utilize the useBytes argument in the grep() function.
By setting useBytes = TRUE, the matching process is carried out byte-by-byte, making the actual matching faster but possibly incompatible with multibyte locales.
# Using useBytes may not be suitable for multibyte characters,
# but can be faster for ASCII-only matching
matches_ascii <- grep("apple", data, perl = TRUE, useBytes = TRUE)
# Displaying the matched elements
matched_elements_ascii <- data[matches_ascii] # Returns "apple"
matched_elements_asciiThe output of the above code is:

2) Working with UTF-8 Strings
When dealing with UTF-8 encoded strings in R, the grep function can be effectively used with the enc2native()function.
The enc2native() function facilitates the conversion of character vectors from various encodings to a native encoding compatible with grep.
Here’s an illustrative example of this approach:
# Sample data with UTF-8 encoded characters
utf8_data <- c("foo", "???", "bar", "???? (abgg)")
# Converting UTF-8 data to native encoding
native_data <- enc2native(utf8_data)
# Searching for a pattern containing UTF-8 characters
matches_utf8 <- grep("???", native_data)
# Displaying the matched elements
matched_elements_utf8 <- native_data[matches_utf8]
matched_elements_utf8The above code works to give the following output:

If you are interested in how R and Power BI can work together, check out our video which further elaborates on the importance of learning R for Power BI users.
Final Thoughts
Grep provides a powerful way to identify matches based on regular expressions or fixed strings. It enables efficient finding of relevant elements even in large datasets.
With robust pattern syntax, grep can handle complex matching needs. Related functions expand capabilities, from logical matches to replacements.
But the relevance of grep in R extends beyond basic pattern matching. It’s a key element in broadening your programming prowess and offers precision and control in your searches.
Whether you are dealing with simple character matching or intricate regular expressions, this tool opens doors to significant data analysis capabilities.
On top of this, the application of these functions is not limited to just R itself. It can also enhance your skills in other data analysis tools such as Power BI. The knowledge you gain here is transferable and broadens your capabilities in data management.
Frequently Asked Questions
Let’s address some additional queries you might have related to the grep function.

1. How does R return matched elements with grep()?
When using grep() in R, the function returns a vector containing the indices of the input vector elements that match the given pattern. To extract the matched elements themselves, you can set the value parameter to TRUE.
For example, see the following R script:
# Create a character vector
words <- c("apple", "banana", "cherry", "date", "fig", "grape")
# Use grep() with a regular expression to find words
# that start with a vowel and end with "e"
grep("^[aeiou].*e$", words, value=TRUE)This gives the following output:

2. Which package contains grepl() function?
Both grep() and grepl() functions are part of R’s base package, which means you can access them without the need to install or load any additional packages.
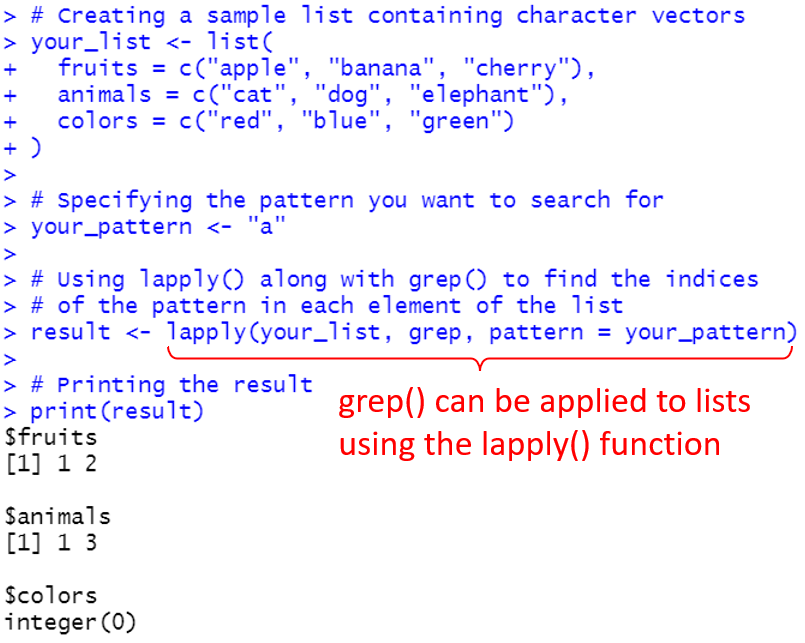
3. How can I use grep on a list in R?
To apply grep() on a list in R, you can use the lapply() function.
This will apply grep() to each element of the list.
For example, lapply(your_list, grep, pattern = “your_pattern”) will return a list of indices for each element in your_list that match the given pattern.
Here’s an example:
# Creating a sample list containing character vectors
your_list <- list(
fruits = c("apple", "banana", "cherry"),
animals = c("cat", "dog", "elephant"),
colors = c("red", "blue", "green")
)
# Specifying the pattern you want to search for
your_pattern <- "a"
# Using lapply() along with grep() to find the indices of the pattern in each element of the list
result <- lapply(your_list, grep, pattern = your_pattern)
# Printing the result
print(result)And, this gives the following output: